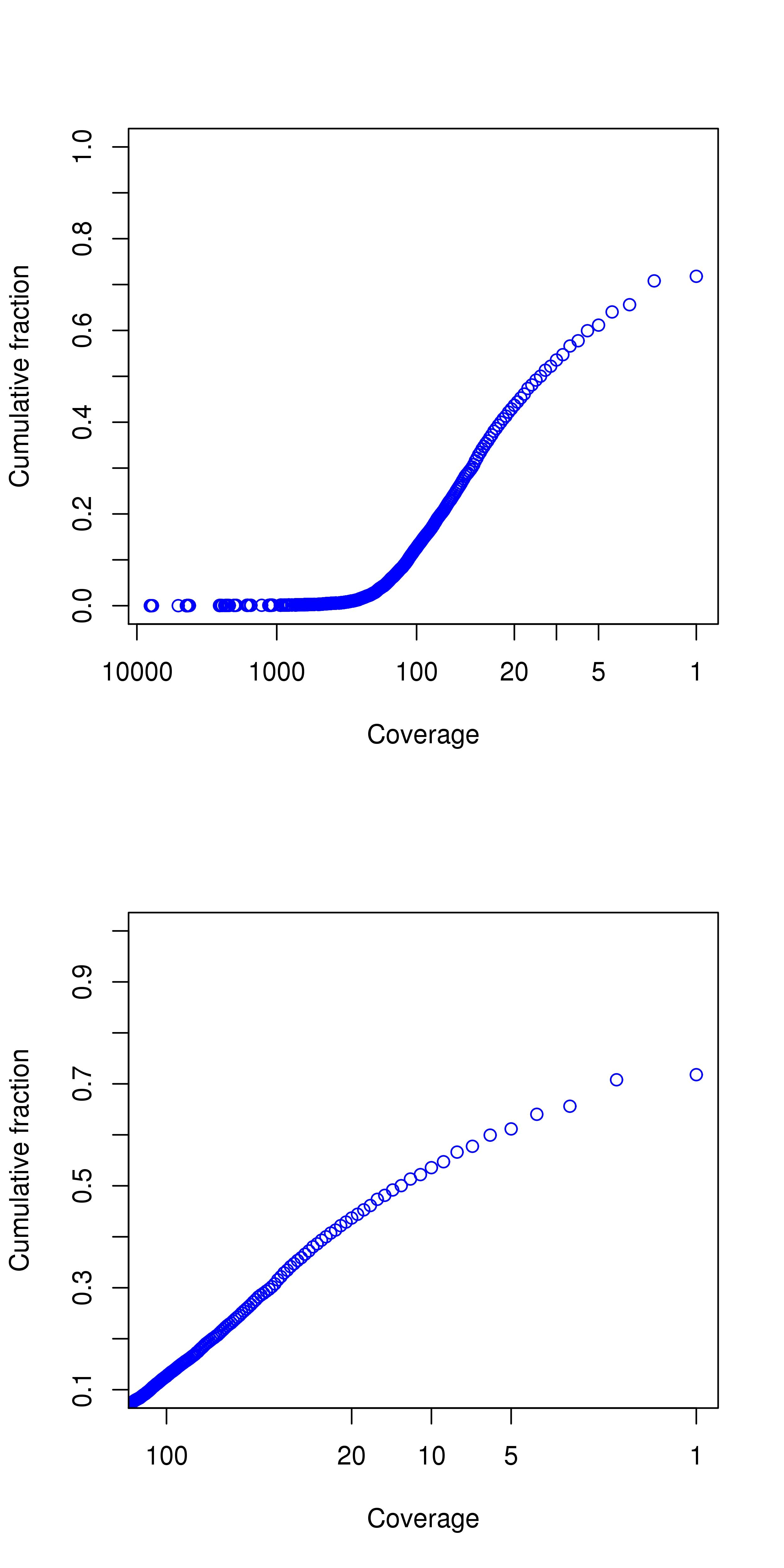
PatientXsomatic
Input Quality Assessment
PatientXsomatic.0.tumor
PatientXsomatic.0
PatientXsomatic.1.tumor
PatientXsomatic.1
Statistics
| source: PatientXsomatic_result/PatientXsomatic.0_bwamem.sort.readfiltered.bam | |
| QC-passed reads | 458754 |
| QC-failed-reads | 0 |
| Duplicates | 0 |
| Mapped reads | 347409(75.73%) |
| Paired reads | 458754 |
| Read1 | 229377 |
| Read2 | 229377 |
| Properly paired | 331618(72.29%) |
| Reads with itself and mate mapped | 347388 |
| Singletons | 21(0.00%) |
| Reads with mate mapped to a different chromosome | 42735 |
| Reads with mate mapped to a different chr (mapQ at least 5) | 42735 |
| source: PatientXsomatic_result/PatientXsomatic.0_bwamem.sort.readfiltered.bam | |
| total length (defined by /auto/rcf-proj/kw/yunfeigu/try_seqmule/somatic/somatic_calling.bed) | 244.59Kb |
| Fraction of reads mapped to target region | 41.59% |
| Average coverage in target region | 42.59 |
| Percentage above 30 | 36.59% |
| Percentage above 20 | 43.68% |
| Percentage above 10 | 53.56% |
| Percentage above 5 | 61.16% |
| source: PatientXsomatic_result/PatientXsomatic.0_bwamem.sort.readfiltered.0_varscan.somatic-call.extract.vcf | |
| Sample | PatientXsomatic |
| Number of variants | 199 |
| Number of SNVs | 178 |
| Number of indels | 21 |
| Transitions | 115 |
| Transversions | 63 |
| Ti/Tv Ratio | 1.83 |
| Ref/Alt heterozygotes | 133 |
| Homozygotes | 66 |
Coverage Plot